Research
Action of proteases in cell death control
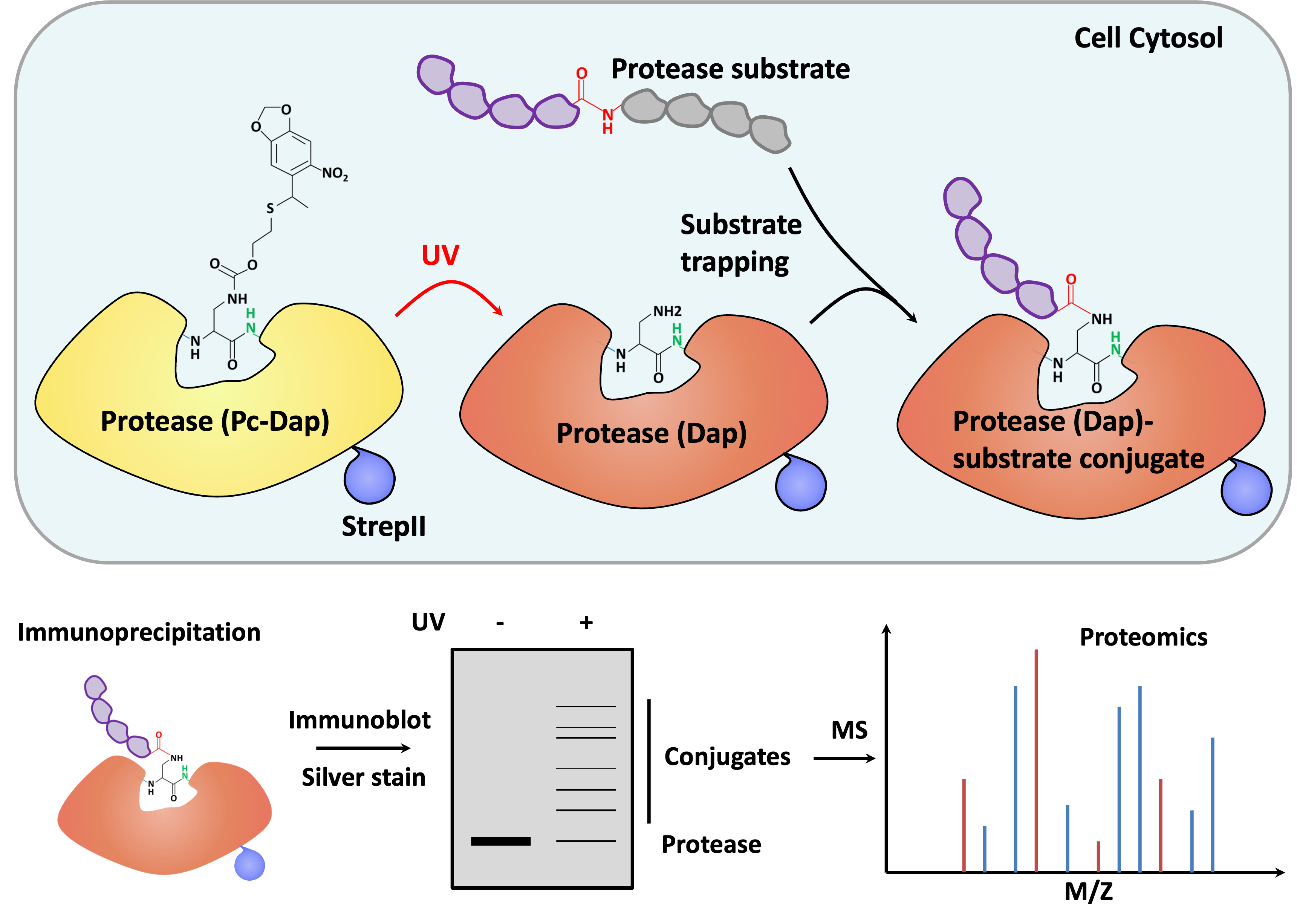
Plants exhibit both similarities and differences with humans in terms of cell death. Unlike humans, plants do not appears undergo apoptosis, but both plants and humans elicit inflammatory cell death in response to pathogen infection: hypersensitive response (HR, plants) and necroptosis/ferroptosis/pyroptosis (human). The mediators and downstream signaling pathways in this process share both similarities and differences between the two systems. In plant, the activation of intracellular nucleotide binding and leucine-rich repeat-containing receptors (NLR), which recognize bacterial effector proteins or effector-mediated changes, leads to the assembly of a supramolecular complex named the resistosome, analogous to human inflammasome. The resistosome can mediate rapid cation influx and trigger HR. Downstream of resistosome activation and the execution of final membrane rupture in plant HR still remains unknown. Several proteases have been reported to play important roles in HR but substrate identification of protease is a challenging question. We used an artificial amino acid engineering and proteomics approach to look for caspase-4 substrates in human and identified over 200 candidate substrates. Here at K-state, we aim to use the same approach to identify critical protease substrates to explore the molecular mechanism of plant HR.
Action of kinases in defense signaling
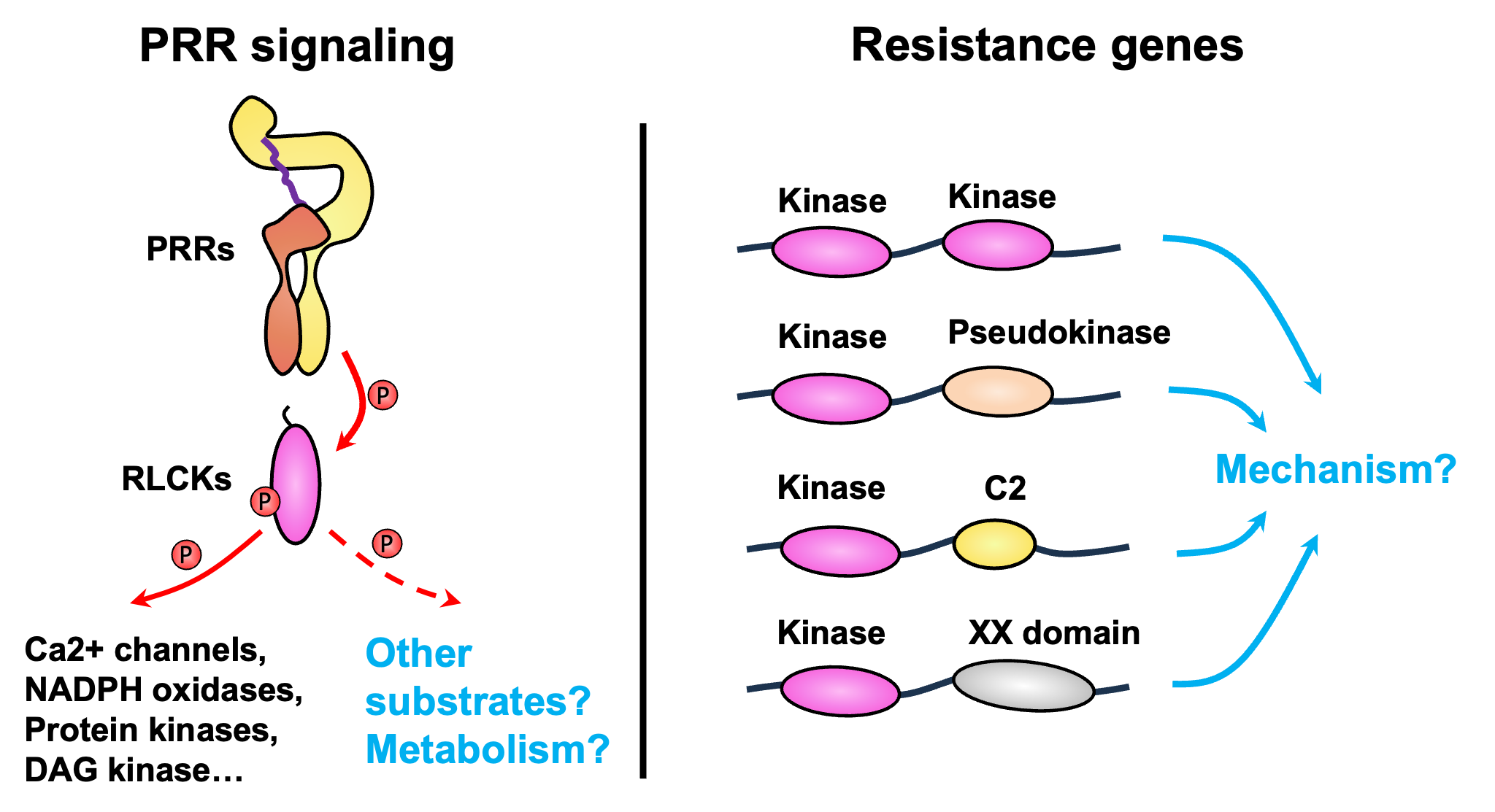
In addition to intracellular receptors such as NLRs, plants rely on plasma membrane–localized pattern recognition receptors (PRRs) to survey the environment and sense pathogens. Activation of multiple PRRs leads to the activation of receptor-like cytoplasmic kinases (RLCKs), which relay signals to downstream components through phosphorylation. Previous research, including our own findings, suggests that RLCKs directly phosphorylate substrates such as ion channels, NADPH oxidases, and lipid kinases to activate defense responses. Do RLCKs phosphorylate other substrates? We have evidence that RLCKs may also be involved in regulating metabolism, which plays an important role in defense responses.
Meanwhile, many resistance genes in different plant species have been identified in recent years that encode novel proteins with kinase domains. How these proteins provide immunity to plants remains unclear. In collaboration with laboratories at Kansas State University, we will study the molecular mechanisms underlying these resistance genes.
Regulation of lipid signaling in plant immunity
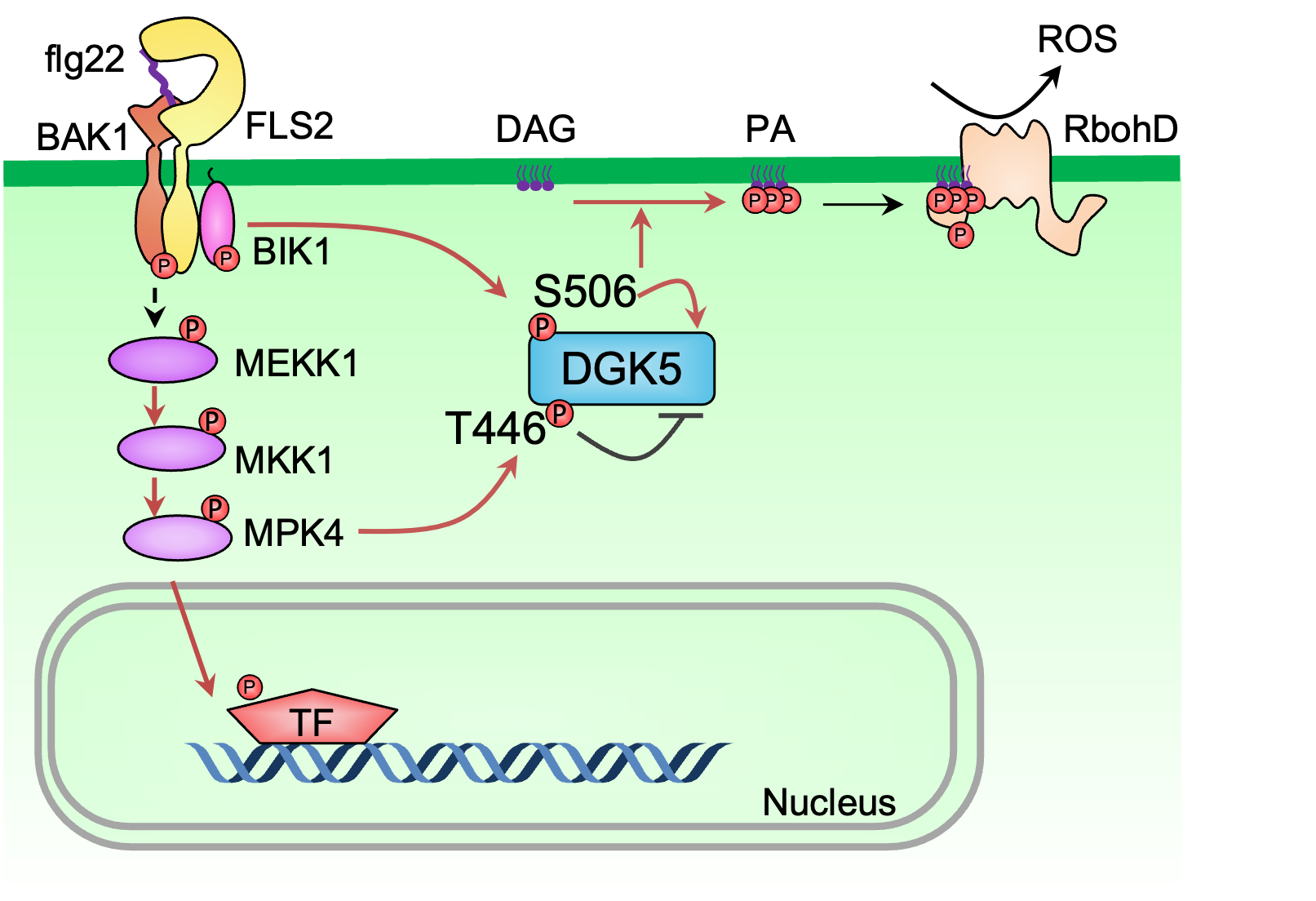
Diacylglycerol kinases (DGKs) phosphorylate diacylglycerol (DAG) to generate phosphatidic acid (PA). PA is a crucial lipid signaling molecule and is rapidly produced upon pathogen perception. Unlike in animal systems, where phospholipase C (PLC) activated by G protein–coupled receptors (GPCRs) initiates lipid signaling, the activation of lipid signaling in plants remained mysterious for decades.
In our 2024 Cell paper (see #10 on the Publications page), we discovered that upon receptor perception of ligand, BIK1 (an RLCK) phosphorylates DGK5 at Ser-506, thereby activating PA production and lipid signaling. Additionally, MPK4 was found to phosphorylate DGK5 at Thr-446. Differential phosphorylation of DGK5 at these distinct sites plays opposing roles in regulating DGK5 activity and PA production, thereby finely tuning plant defense responses.
Our work, for the first time, established a direct link between innate immunity and lipid signaling in plants and resolved a long-standing mystery regarding how immune activation triggers lipid signaling. However, many questions remain regarding the regulation of lipid signaling in plant immunity, and we are actively working to address them.
Engineering of disease resistant plants
Understanding above research questions will not only deepen our knowledge of the molecular mechanisms underlying plant immunity, but also provide valuable foundations for translational applications. By combining mechanistic insights with innovative molecular engineering approaches, we aim to develop strategies for creating crops with broad-spectrum, durable resistance to pathogens.